Testing Charm Biotech DNA Extraction on Cryosectioned P. comp and M. cap
Testing Charm DNA Kit on P. comp and M. cap
Protocol Link
Sample prep notes for extracting from whole sections instead of LCM samples:
- Remove a cryosectioned slide from -80 ºC (same sample/sectioning round as used for LCM), transfer to -20 ºC for 30 minutes, then transfer slide to cooler with dry ice
- KEEP CONTAINER CLOSED, with dessicant inside
- “take slide in container with desiccant out of -80 ºC and allow to slowly reach room temperature, for at least 60 min, before opening (Roy et al 2020 Bioprotocols)”
- KEEP CONTAINER CLOSED, with dessicant inside
- Pre-cool lysis buffer mixture on ice
- Pre-cool forceps and razor blades for transferring tissue to lysis buffer
- Stain and wash all slides and perform dissections in petri dishes cleaned to be RNAse free
- Remove OCT, in RNAse free petri dish on ice, pipette solutions on and off slide with 1mL pipette
- 2 minute in ice-cold 70% Ethanol
- 45 s in ice-cold 50% Ethanol
- 30 s in ice-cold 30% Ethanol
- if there is excess OCT, dip slide in ice-cold RNAse free water 5-6 times over 1-2 mins
- Remove slide to dark background over PCR rack (over dry ice, though be careful to not freeze the water once the volume on the slide is low, if that’s freezing move to regular ice) and use sterile razor blade, cleaned with RNAse cleaner, to scrape off tissue sections into a tube filled with prepared 100 uL digestion buffer.
Testing two versions of the protocol
- LCM FFPE protocol as written, without the initial cap 60 ºC upside down incubation (cells already in bottom of tube) and without the 94 ºC incubation, since this seems too high for our non-FFPE samples. 60 ºC hour incubation in thermomixer, with no shaking, for 3 hours. Eluted in 20 uL elution buffer.
- Same as above, but based on the RNA extraction results, shortening the incubation to do what the Qiagen PAXgene kits do: shaker–incubator for 1 h at 56°C and 1400 rpm.
Samples:
- P_1 - POR DD2 buffer 1 hr incubation, 56 ºC, 1400rpm
- P_3 - POR DD2 buffer 3 hr incubation, 60 ºC (written FFPE protocol, minus the 94 ºC step)
- M_1 - MON DD2 buffer 1 hr incubation, 56 ºC, 1400rpm
- M_3 - MON DD2 buffer 3 hr incubation, 60 ºC (written FFPE protocol, minus the 94 ºC step)
Notes
Notes: both of the Porites eluted DNA were foamy at the end of the elution…
Quality Control (QC)
These steps analyze the quantity and quality of the DNA extracted.
DNA Quantity
Follow Broad Range dsDNA Qubit protocol to analyze sample quantity. Read standards once and record values, read all samples twice.
DNA Standards: 414.64 (S1) & 8566.59 (S2)
| sample_id | Species | DNA_QBIT_1 | DNA_QBIT_2 | DNA_QBIT_AVG |
|---|---|---|---|---|
| P_1 - POR 1 hr, 56 ºC, 1400rpm | Porites compressa | 21.8 | 21.8 | 21.8 |
| P_3 - POR 3 hr, 60 ºC | Porites compressa | 46.4 | 46.6 | 46.5 |
| M_1 - POR 1 hr, 56 ºC, 1400rpm | Montipora capitata | 13.0 | 13.0 | 13.0 |
| M_3 - POR 3 hr, 60 ºC | Montipora capitata | 13.0 | 12.7 | 12.9 |
DNA Quality
If DNA quantity is sufficient (typically >10 ng/µL) follow the PPP Agarose Gel Protocol to determine DNA quality. “Good” DNA should form a distinct band a the very top of the gel.
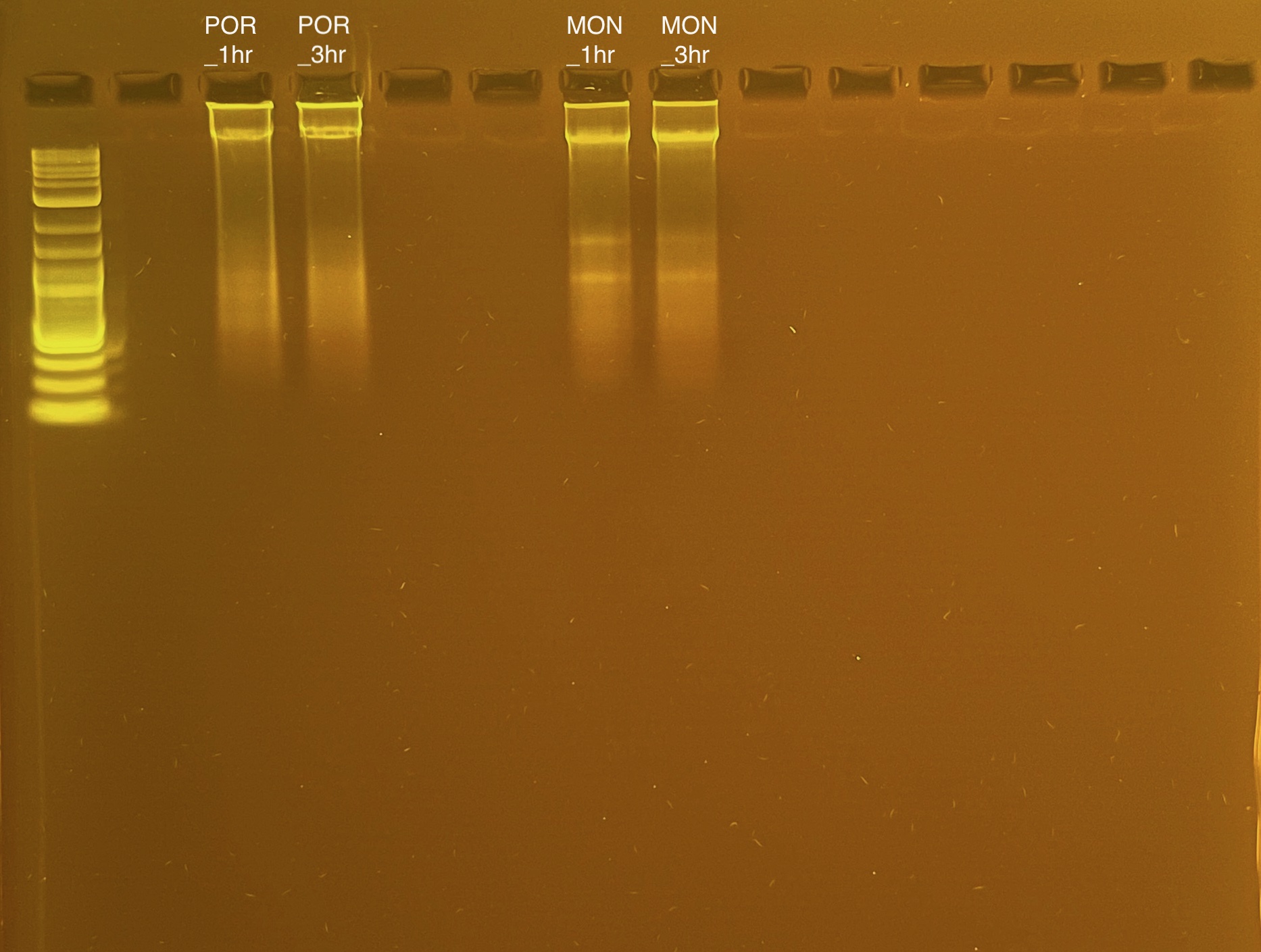
Written on February 9, 2024