SwitchFree library prep
“TagSeq” Zymo Switch-Free RNA Library Prep Test/Protocol
Using product: Zymo-Seq SwitchFree 3’ mRNA Library Kit. See Zymo’s protocol here. Basing this protocol directly off of Jill Ashey’s successful testing of the kit for Pocillopora larval samples.
Goal for today
Today, Natalie and I will test the library prep on three adult samples from my Time Series project. We will do all three with two versions of the protocol: one per sample as written and one prep per sample with 3 uL of Poly A reagent instead of 5 uL.
The kit needs a minimum of 10 ng of total RNA or a maximum of 500 ng of total RNA. We will be using 20 ng of total RNA as input. Here’s a breakdown of input RNA volumes for each sample:
| TubeID | Qubit RNA (ng/uL) | Strip tube # | RNA (uL) | RNAse/DNase-Free water (uL) | Total starting volume (ul) | PolyA R1 volume (ul) |
|---|---|---|---|---|---|---|
| POC_R12_C3 | 28.6 | 1 | 0.70 | 4.30 | 5.0 | 5.0 |
| POC_R1_H1 | 24.6 | 2 | 0.81 | 4.19 | 5.0 | 5.0 |
| POC_R3_C3 | 17.7 | 3 | 1.13 | 3.87 | 5.0 | 5.0 |
| POC_R12_C3 | 28.6 | 4 | 0.70 | 4.30 | 5.0 | 3.0 |
| POC_R1_H1 | 24.6 | 5 | 0.81 | 4.19 | 5.0 | 3.0 |
| POC_R3_C3 | 17.7 | 6 | 1.13 | 3.87 | 5.0 | 3.0 |
Here’s the SwitchFree library prep workflow:
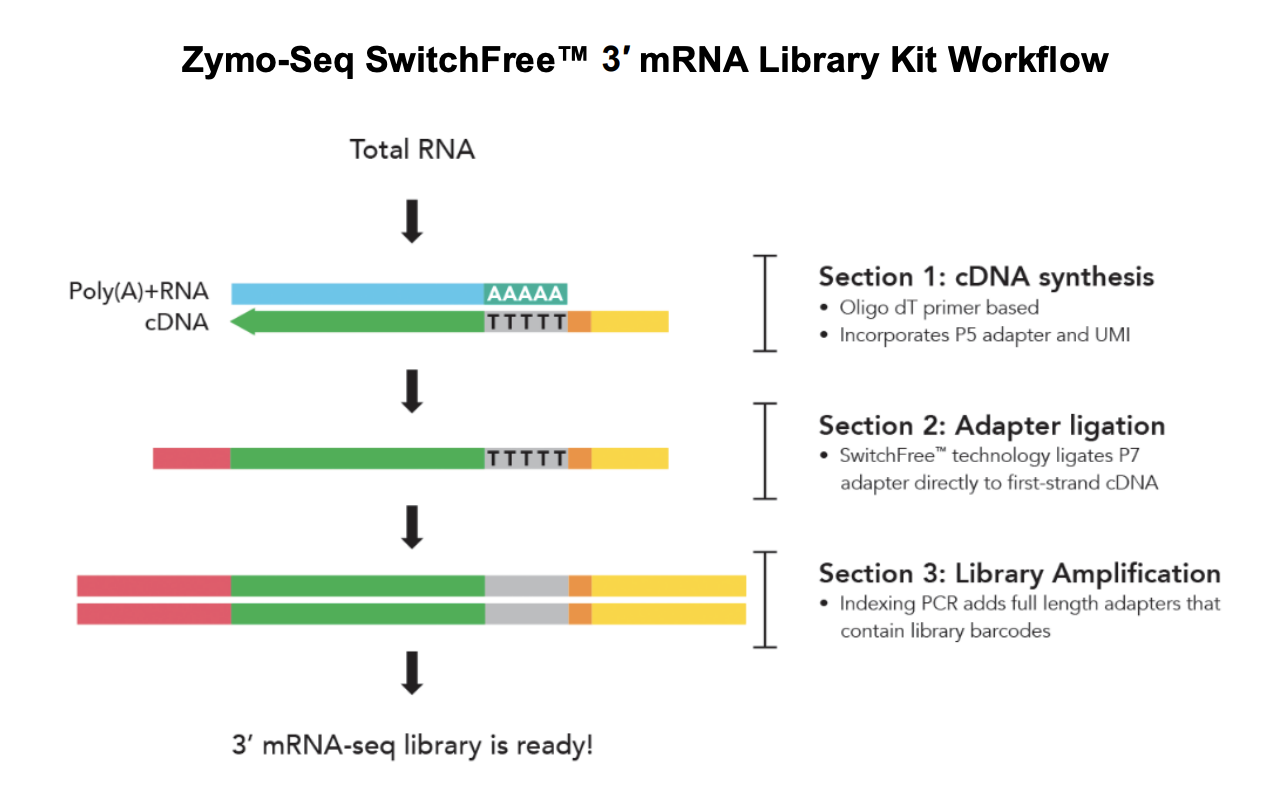
Materials
- Zymo-Seq SwitchFree 3’ mRNA Library Kit
- PCR tubes
- Thermocycler
- Mini centrifuge
- Aluminum beads (to keep things on ice)
- Magnetic stand for PCR tubes
- Kit contents
Buffer preperation
Once buffers are prepared for a kit, they do not need to be prepared again.
- Add 300 uL of the Select-a-Size MagBead concentrate to 10 mL of Select-a-Size MagBead Buffer.
- These should be prepared at least 5 days ahead of library prep.
- Add 24 mL of 100% ethanol to the DNA Wash Buffer concentrate. Store at room temperature.
Best practices
- Avoid multiple freeze thaws of all components, and aliquot components as necessary
- Remove enzymes from cold storage just before use and return to cold storage immediately after use
- Thaw and maintain all components on ice unless noted otherwise
- Flick to mix thawed components and briefly centrifuge prior to use
- After adding each component, mix by pipetting up and down 15-20 times. Briefly centrifuge after
- Pre-program thermal cycler with lid heating ON set to >100-105°C
- Turn on thermocycler day-of to preheat
- Pre-calculate how much to dilute RNA samples
Protocol
Section 1: cDNA Synthesis
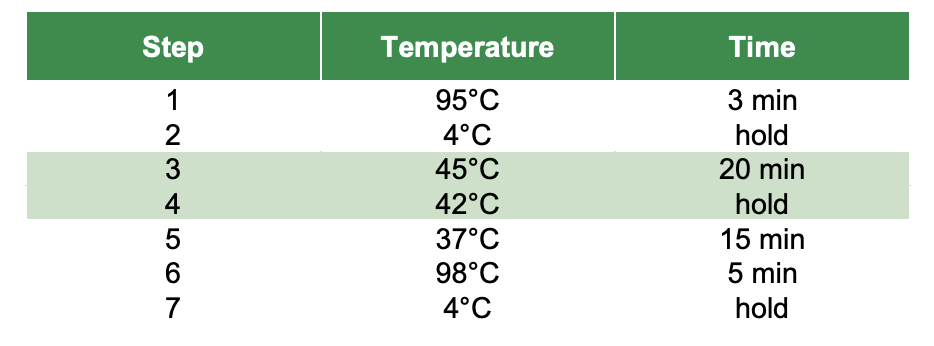
Thermocycler settings:
- Thaw PolyA R1 Reagent and R2 Reagent on ice
- Prepare samples to correct volume in strip tubes
- Add 5 uL of PolyA R1 Reagent to each sample
- AS A TEST: FOR HALF OF SAMPLES, ADD 3 uL PolyA R1 and 2 uL RNAse/DNAse-free water
- Mix by pipetting and centrifuge briefly
- Run Steps 1-2 in thermocycler
- SKIP this step if RNA is degraded/low quality
- While in the thermocycler on the 4°C hold, add 10 uL of R2 Reagent to each sample
- Mix by pipetting
- Run Steps 3-4
- While Steps 3-4 are running, allow Reaction Clean-up Solution to equilibrate to room temperature
- While in the thermocycler on the 42°C hold, add 4 uL of Reaction Clean-up Solution to each sample
- Mix by pipetting and centrifuge briefly
- Immediately return to thermocycler
- Run Steps 5-7
- Remove tubes from thermocycler
- Add 26 uL of 95% ethanol to each sample
- Do bead clean-up with Select-a-Size MagBeads
- Allow beads to equilibrate to room temperature >30 minutes before use
- Resuspend beads immediately before use by shaking or vortexing until homogenous
- Add 110 uL of Select-a-Size MagBeads to each sample
- Pipette to mix until homogenous
- Incubate for 5 minutes at room temperature
- Put samples on magnetic stand for 5 mins or until beads have fully separated from solution
- Aspirate slowly and discard supernatent
- While the sample is still on the magnetic stand, add 200 uL of DNA Wash Buffer without disturbing the pellet
- Aspirate slowly and discard supernatent
- Repeat this wash step again
- Keep the caps open to air-dry bead for 1 minute
- Aspirate any residual wash buffer
- Continue to air dry pellet until it appears matte without cracking (see picture below)
- Remove sample from magnetic stand
- Add 10 uL of DNA Elution Buffer to each sample
- Pipette to mix until homogenous
- Put samples on magnetic stand for 1-2 minutes or until elute is clear
- Move elute to new PCR tubes
Section 2: Adapter Ligation
Thermocycler settings:
Step 1: 95°C for 3 minutes, 4°C for 2 minutes, 4°C hold
Step 2: 25°C for 45 minutes (lid temperature at 40°C), 4°C hold
- Thaw 3’mRNA L Reagent on ice
- Put samples in thermocycler and run Step 1
- Remove samples from thermocycler and put on ice
- Add 15 uL of 3’mRNA L Reagent to each sample
- Mix by pipetting and centrifuge briefly
- Run Step 2
- Remove samples from thermocycler
- Add 25 uL of DNase/RNase free water to each sample
- Do bead clean-up with Select-a-Size MagBeads
- Allow beads to equilibrate to room temperature >30 minutes before use
- Resuspend beads immediately before use by shaking or vortexing until homogenous
- Add 100 uL of Select-a-Size MagBeads to each sample
- Pipette to mix until homogenous
- Incubate for 5 minutes at room temperature
- Put samples on magnetic stand for 5 mins or until beads have fully separated from solution
- Aspirate slowly and discard supernatent
- While the sample is still on the magnetic stand, add 200 uL of DNA Wash Buffer without disturbing the pellet
- Aspirate slowly and discard supernatent
- Repeat this wash step again
- Keep the caps open to air-dry bead for 1 minute
- Aspirate any residual wash buffer
- Continue to air dry pellet until it appears matte without cracking (see picture above)
- Remove sample from magnetic stand
- Add 15 uL of DNA Elution Buffer to each sample
- Pipette to mix until homogenous
- Put samples on magnetic stand for 1-2 minutes or until elute is clear
- Move elute to new PCR tubes
Section 3: Library amplification
Thermocycler settings:
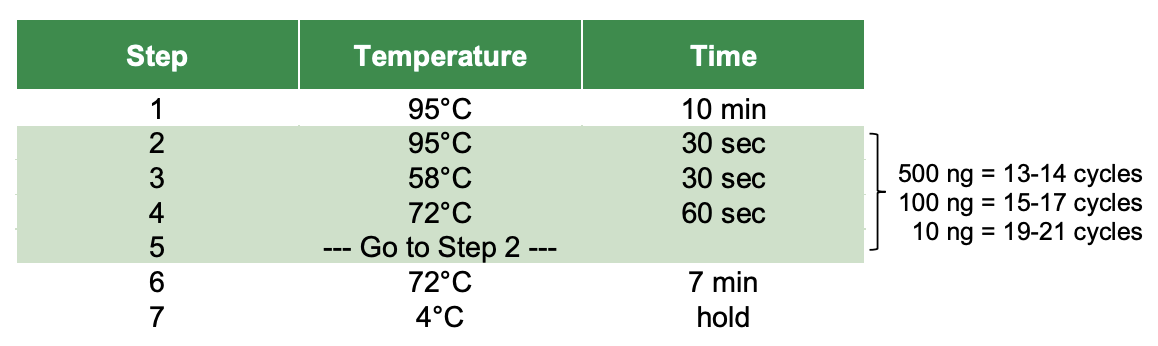
Given the low input for these preps and my previous experience with other Zymo kits, we are going to do 21 PCR amplification cycles.
- Thaw Index Primers and ZymoTaq Premix on ice
- Allow Library Binding Solution to equilibrate to room temperature >30 minutes before use
- Add 10 uL of unique Index Primer to each sample
- Mix by pipetting
- Add 25 uL of Zymo TaqPremix to each sample
- Mix by pipetting and centrifuge briefly
- Run thermocycler program above
- Remove samples from thermocycler
- Add 50 uL of DNase/RNase free water to each sample
- Add 80 uL of Select-a-Size MagBeads to each sample
- Pipette until homogenous
- Incubate for 5 minutes at room temperature
- Place samples on magnetic stand for 5 minutes or until beads have separated from solution
- Aspirate slowly and discard supernatent
- Remove samples from magnetic stand
- Add 100 uL of DNA Elution Buffer to beads
- Pipette until homogenous
- Add 80 uL of Library Binding Solution to each sample
- Pipette until homogenous
- Incubate for 5 minutes at room temperature
- Do bead clean-up with Select-a-Size MagBeads (beads already added to samples)
- Put samples on magnetic stand for 5 mins or until beads have fully separated from solution
- Aspirate slowly and discard supernatent
- While the sample is still on the magnetic stand, add 200 uL of DNA Wash Buffer without disturbing the pellet
- Aspirate slowly and discard supernatent
- Repeat this wash step again
- Keep the caps open to air-dry bead for 1 minute
- Aspirate any residual wash buffer
- Continue to air dry pellet until it appears matte without cracking (see picture above)
- Remove sample from magnetic stand
- Add 15-20 uL of DNA Elution Buffer to each sample
- Pipette to mix until homogenous
- Put samples on magnetic stand for 1-2 minutes or until elute is clear
- Move elute to new PCR tubes
THIS IS THE FINAL 3’ mRNA-seq LIBRARY. STORE AT -20°C.
QC
- Used Broad range DNA Qubit Protocol
- All samples read twice, standard only read once
DNA Standards: 182.07 (S1) & 23026.07 (S2)
| TubeID | PolyA R1 volume (ul) | UDI | Library_QBIT_1 | Library_QBIT_2 | Library_QBIT_AVG |
|---|---|---|---|---|---|
| POC_R12_C3 | 5.0 | UDI01 | 4.14 | 4.10 | 4.12 |
| POC_R1_H1 | 5.0 | UDI02 | 3.22 | 3.16 | 3.19 |
| POC_R3_C3 | 5.0 | UDI03 | 3.94 | 4.00 | 3.96 |
| POC_R12_C3 | 3.0 | UDI08 | 6.00 | 6.00 | 6.00 |
| POC_R1_H1 | 3.0 | UDI07 | 3.62 | 3.60 | 3.61 |
| POC_R3_C3 | 3.0 | UDI06 | 4.96 | 5.02 | 4.98 |
Tapestation
Run DNA Tapestation to visualize libraries. Here’s an example of what the library should look like on a Tapestation:
EXAMPLE:
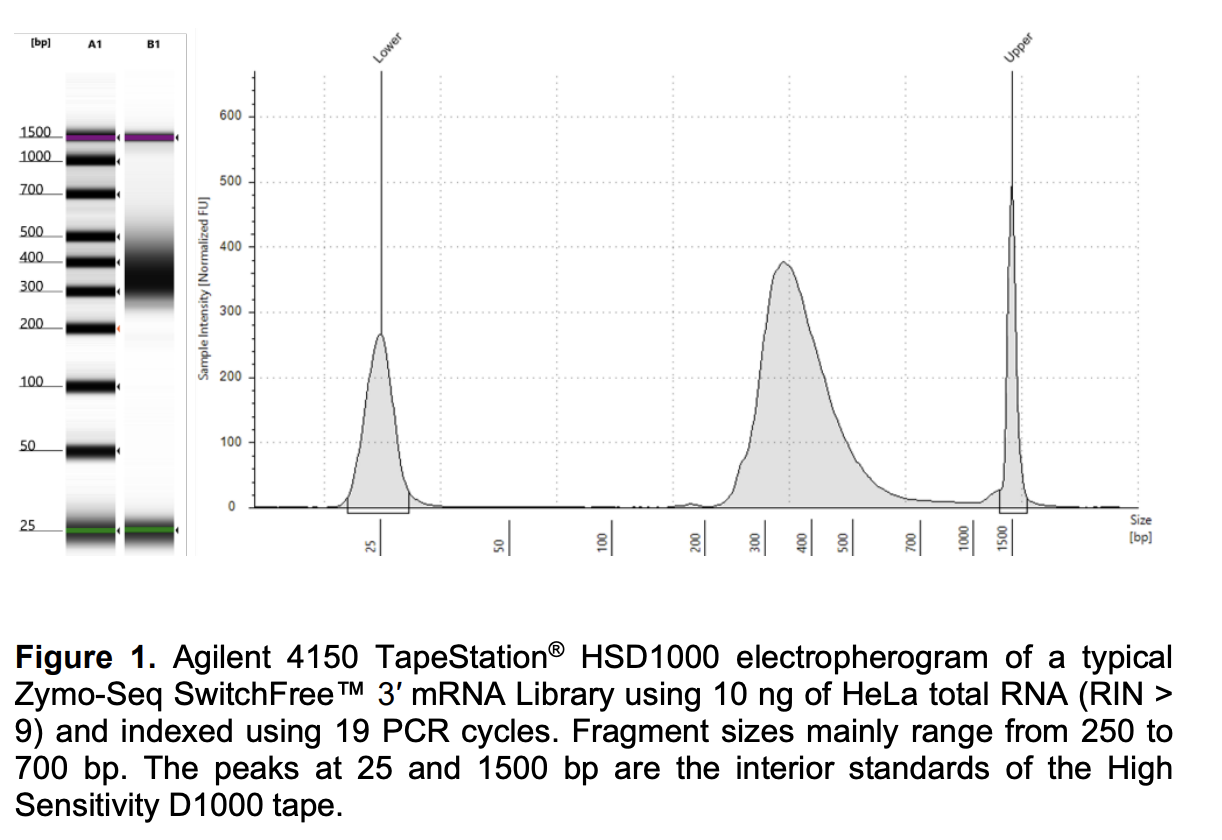


| TubeID | 5 ul PolyA reagent | 3 ul PolyA |
|---|---|---|
| POC_R12_C3 |  |
 |
| POC_R1_H1 |  |
 |
| POC_R3_C3 |  |
 |
Full results can be found here and here (not scaled by sample).
Thoughts
- Did 21 cycles when I meant to do 19 - could be the cause of the dimers. However, concentration is lower than ideal. I wonder if increasing or decreasing the RNA input amount could help…
Do we have enough to sequence? Let’s Convert library concentration from ng/μL to nM
First, library concentrations were converted from ng/μL to nM using the equation:
(concentration in ng/μL)/(660g/mol * average library size in bp) * 10^6 = concentration in nM

| Library_Tube | Library_QBIT_1 | Library_QBIT_2 | UDI | TubeID | PolyA R1 volume (ul) | Qubit_Conc | TS_Conc | TS_Peak_Size | Molarity_nM_Qubit | Molarity_nM_TS_Calculated | Molarity_nM_TS_Peak |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4.14 | 4.1 | UDI01 | POC_R12_C3 | 5 | 4.12 | 4.63 | 381 | 16.38 | 18.41 | 17.50 |
| 2 | 3.22 | 3.16 | UDI02 | POC_R1_H1 | 5 | 3.19 | 4.7 | 406 | 12.69 | 18.69 | 17.10 |
| 3 | 3.94 | 4 | UDI03 | POC_R3_C3 | 5 | 3.96 | 4.72 | 408 | 15.75 | 18.77 | 16.90 |
| 4 | 6 | 6 | UDI08 | POC_R12_C3 | 3 | 6 | 6.31 | 399 | 23.86 | 25.09 | 24.30 |
| 5 | 3.62 | 3.6 | UDI07 | POC_R1_H1 | 3 | 3.61 | 4.32 | 407 | 14.36 | 17.18 | 16.30 |
| 6 | 4.96 | 5.02 | UDI06 | POC_R3_C3 | 3 | 4.98 | 5.23 | 409 | 19.80 | 20.80 | 19.20 |
If we are okay with the dimers, we would now be able to normalize and pool for sequencing
- Normalize library: Dilute to 4nM in a volume of 6-10μL with molecular grade water using the calculation C1V1=C2V2.
- Combined into pool (n=XX libraries per pool) for sequencing
- Quantified with Qubit dsDNA HS Assay Kit (Invitrogen #Q33231) and tapestation analysis with the D5000 ScreenTape
| Library_Tube | Amount_Library_4nM | Amount_H20_4nM |
|---|---|---|
| 1 | 2.44 | 7.56 |
| 2 | 3.15 | 6.85 |
| 3 | 2.54 | 7.46 |
| 4 | 1.68 | 8.32 |
| 5 | 2.79 | 7.21 |
| 6 | 2.02 | 7.98 |
I probably could normalize the concentration of all the libraries and then do a pooled cleanup, or clean them all up again and then requantify and then normalize and pool.
Cleanup to get rid of dimers: Appendix C of protocol
“The kit is designed to minimize the formation of adapter dimers. If significant dimers show up at ~ 180 bp on the size distribution profile, a bead clean-up is recommended to remove the dimers for better sequencing quality. To remove the dimers, use DNase/RNase-Free Water to raise the library volume to 100 µL. Then follow the clean-up protocol in Appendix A, page 10 using 85 µL of the Select-a-Size MagBeads and elute with 15 – 20 µL of DNA Elution Buffer.”
Protocol: 0.85X cleanup
- Add 87 uL of DNase/RNase free water to each thawed library (100 - 13 uL = 87 uL)
- Add 85 uL of Select-a-Size MagBeads to each sample
- Pipette until homogenous
- Incubate for 5 minutes at room temperature
- Place samples on magnetic stand for 5 minutes or until beads have separated from solution
- Aspirate slowly and discard supernatent
- While the sample is still on the magnetic stand, add 200 uL of DNA Wash Buffer without disturbing the pellet
- Aspirate slowly and discard supernatent
- Repeat this wash step again
- Keep the caps open to air-dry bead for 1 minute
- Aspirate any residual wash buffer
- Continue to air dry pellet until it appears matte without cracking (see picture above)
- Remove sample from magnetic stand
- Add 20 uL of DNA Elution Buffer to each sample
- Pipette to mix until homogenous
- Put samples on magnetic stand for 1-2 minutes or until elute is clear
- Move elute to new PCR tubes
THIS IS THE FINAL 3’ mRNA-seq LIBRARY. STORE AT -20°C.
QC of dimer-cleaned libraries
Run DNA Tapestation to visualize libraries.
- Used Broad range (or high sensitivity) DNA Qubit Protocol
- All samples read twice, standard only read once
BR DNA Standards: 185.08 (S1) & 23129.30 (S2) HS DNA Standards (results in bold): 49.32 (S1) & 23034.76 (S2)
| TubeID | PolyA R1 volume (ul) | Tube # | Library_QBIT_1 | Library_QBIT_2 | Library_QBIT_AVG |
|---|---|---|---|---|---|
| POC_R12_C3 | 5.0 | 1 | 2.28 | 2.24 | 2.26 |
| POC_R1_H1 | 5.0 | 2 | 1.26 | 1.24 | 1.25 |
| POC_R3_C3 | 5.0 | 3 | 1.38 | 1.37 | 1.38 |
| POC_R3_C3 | 3.0 | 6 | 2.62 | 2.56 | 2.59 |

Full results can be found here.
Next Step: Equilize all libraries to 4nM concentration and pool
| PostClean_Qubit_Conc | PostClean_TS_Conc | PostClean_TS_Peak_Size | PostClean_Molarity_nM_Qubit | PostClean_Molarity_nM_TS_Calculated | PostClean_Molarity_nM_TS_Peak | PostClean_Amount_Library_4nM | PostClean_Amount_H20_4nM |
|---|---|---|---|---|---|---|---|
| 2.26 | 2.29 | 427 | 8.02 | 8.13 | 8.10 | 4.99 | 5.01 |
| 1.25 | 1.52 | 441 | 4.29 | 5.22 | 5.31 | 9.31 | 0.69 |
| 1.38 | 1.93 | 454 | 4.61 | 6.44 | 6.23 | 8.69 | 1.31 |
| 6 | 6.31 | 399 | 22.78 | 23.96 | 24.30 | 1.76 | 8.24 |
| 3.61 | 4.32 | 407 | 13.44 | 16.08 | 16.30 | 2.98 | 7.02 |
| 2.59 | 2.58 | 471 | 8.33 | 8.30 | 8.41 | 4.80 | 5.20 |
- For each library, added calcucated amount of DNA elution buffer (“PostClean_Amount_H20_4nM”) and the library (“PostClean_Amount_Library_4nM”) to a new PCR tube and mixed well for a total of 10 uL volume for each normalized library.
- Then combined all of these tubes into one LoBind 1.5 mL tube and QC’d. Then froze overnight for shipping next day.
Final pool, sent on 8/19/25 to Oklahoma
| Pool_Qubit_Conc | Pool_TS_Conc | Pool_TS_Peak_Size | Pool_Molarity_nM_Qubit | Pool_Molarity_nM_Tapestation |
|---|---|---|---|---|
| 1.02 | 1.2 | 440 | 3.51 | 4.19 |


Full results can be found here.
Sequencing plan:
Next-Generation Sequencing
- Instrument: Illumina NovaSeq X Plus - 25B - PE 150 Cycle
- Read length: 2 x 150bp (Paired End)
- Pricing unit: per lane
- Number of samples: 1 pool (6 libraries)
- Guaranteed total number of pass filter PE reads/sample: 60M (30M in each direction)
- Deliverables: FastQ files uploaded to Genohub project bucket
up to 99 Mreads for the cost of $3.87 per Mread. Your quote will be adjusted to account for the pricing as per Mread rather than per lane.
