Testing Charm Biotech RNA Extraction on Cryosectioned and LCM-d P. acuta
Testing Charm RNA Kit on LCM Samples
Protocol Link
5/9/24 - Extracting RNA from LCM-captured cells (LCM date 5/8/24)
Used Charm Biotech Just-a-Tube ™ Laser Captured Microdissection (LCM)Sample Total RNA/MicroRNA Purification Kit, protocol
Sectioning performed on 5/6/24 and LCM on 5/9/24. See details here
Protocol notes:
- Followed the Charm FFPE protocol almost exactly, but did all incubations at 56 ºC based on other Paxgene extraction protocols. Did a 3 hour incubation in the thermoblock.
Qubit Results
- Used High Sensitivity RNA Qubit Protocol
- All samples read twice, standard only read once
RNA Standards: 27.24 (S1) & 325.15 (S2)
| colony_id | RNA_QBIT_1 | RNA_QBIT_2 | RNA_QBIT_AVG |
|---|---|---|---|
| LCM sample #3 | nd | nd | nd |
| LCM sample #10 | nd | nd | nd |
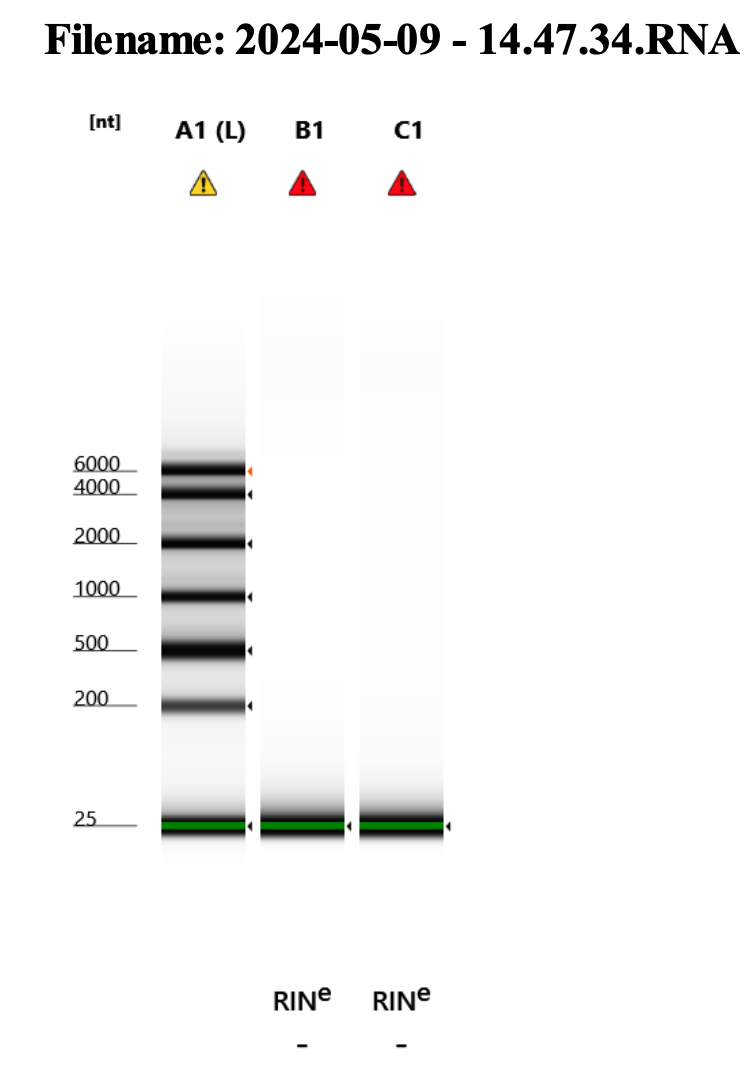
RNA Quality Check: Tapestation
Basically nothing! Oh well.
Full results can be found here
5/13/24 - Attempt #2 of Extracting RNA from LCM-captured cells (LCM date 5/8/24)
Protocol notes:
- Followed the Charm FFPE protocol almost exactly, but did all incubations at 56 ºC based on other Paxgene extraction protocols. Did a 15 minute incubation in the incubator genie.
Qubit Results
- Used High Sensitivity RNA Qubit Protocol
- All samples read twice, standard only read once
RNA Standards: 27.80 (S1) & 310.89 (S2)
| colony_id | RNA_QBIT_1 | RNA_QBIT_2 | RNA_QBIT_AVG |
|---|---|---|---|
| LCM sample #2 | nd | nd | nd |
| LCM sample #6 | nd | nd | nd |
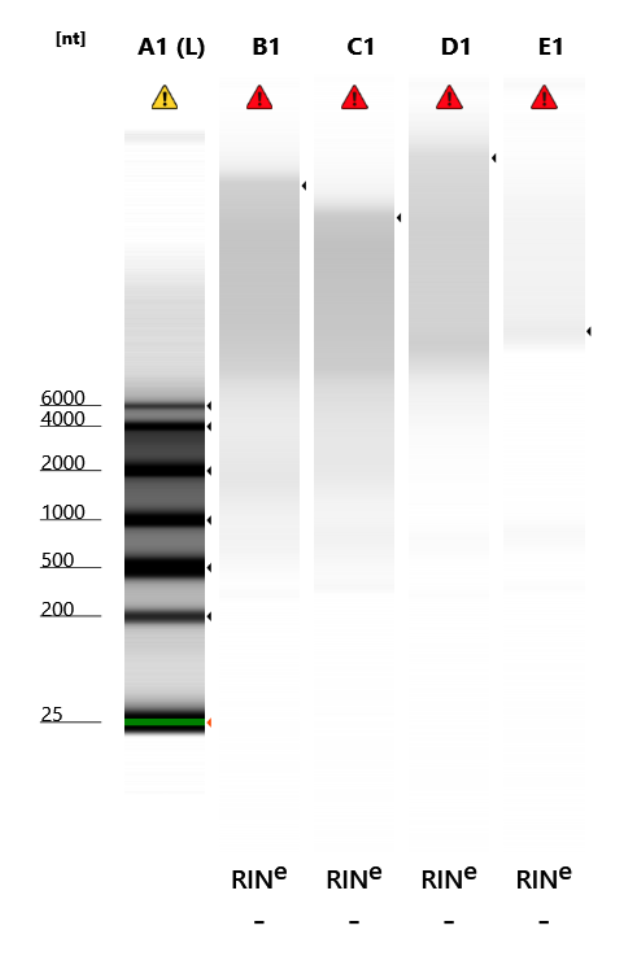
RNA Quality Check: Tapestation
Ran a high sensitivity RNA tapestation for the first time. Is this degraded DNA? Got RINs and concentrations but this doesn’t look like RNA.
Full results can be found here
What do I get if I do a HS ds Qubit on these? Are they DNA?
Qubit Results
- Used High Sensitivity DNA Qubit Protocol
- All samples read twice, standard only read once
DNA Standards: 67.25 (S1) & 22077.28 (S2)
| colony_id | DNA_QBIT_1 | DNA_QBIT_2 | DNA_QBIT_AVG |
|---|---|---|---|
| LCM sample #3 | 0.913 | 0.924 | 0.9185 |
| LCM sample #10 | 0.793 | 0.798 | 0.7955 |
| LCM sample #2 | 1.32 | 1.35 | 1.335 |
| LCM sample #6 | 1.71 | 1.68 | 1.695 |
So, there is DNA!
DNAse I digestion
https://www.zymoresearch.com/products/dnase-i-set
https://files.zymoresearch.com/protocols/_r1013_r1014_r1015_r1016_rna_clean_concentrator-5.pdf
Sample final volume is 13 uL. Added 1.875 uL of DNAse digestion buffer and 1.875 uL of DNAse. Incubated 15 mins at room temp.
- Heat to 75ºC for 15 mins to inactivate the DNAse. Should I just skip this step and run a tapestation right away? Seems like it will just degrade the RNA more.
- Skipping this for today, since we know we will not use this RNA for any cDNA synthesis. in the future, definitley deactivate the DNAse.
RNA Quality Check: Tapestation
What does the RNA look like after DNAse digestion?
Not great! Missing the lower marker? Any chance this is because of the DNAse? Either way, when blowing up the contrast I can see bands that could (?) be RNA but honestly it looks unlikely.
Full results can be found here