2023-04-11 E5 Sites 2 & 3 Species ID (POC and POR) via PCR and Sanger Sequencing
E5 Species ID (POC and POR) via PCR and Sanger Sequencing: Sites 2 and 3
Link with sample list and metadata
See base protocol and notes from Hollie
Followed my PCR Protocol exactly, based on instruction from Kristina Terpis
Sample List: All POC and POR colonies from the E5 timeseries need to be ID’d for one timepoint per colony. Starting with Site 2 and 3. Almost all samples are from Time Point 2 unless otherwise noted in the metadata (because no molecular sample was taken at TP2).
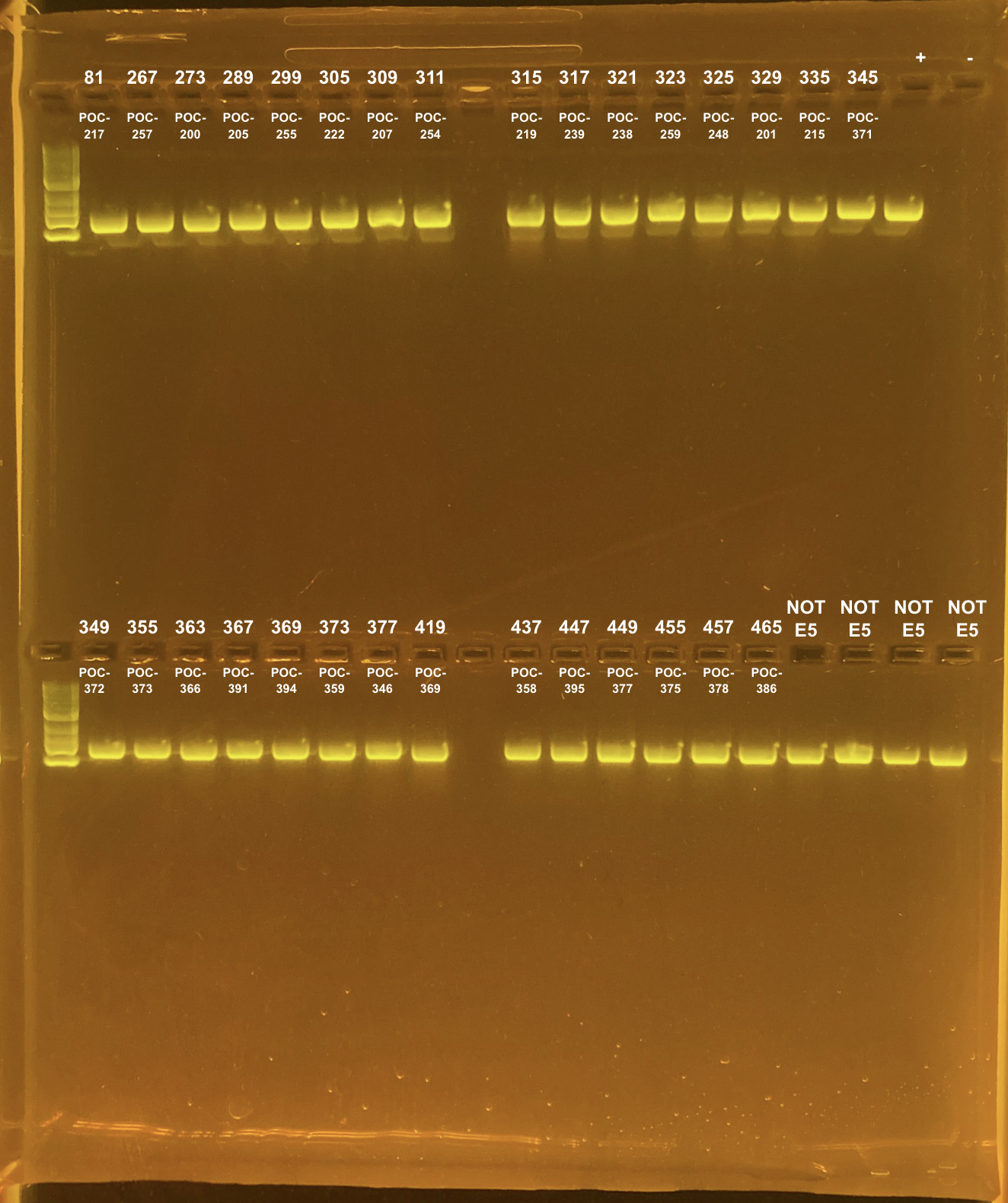
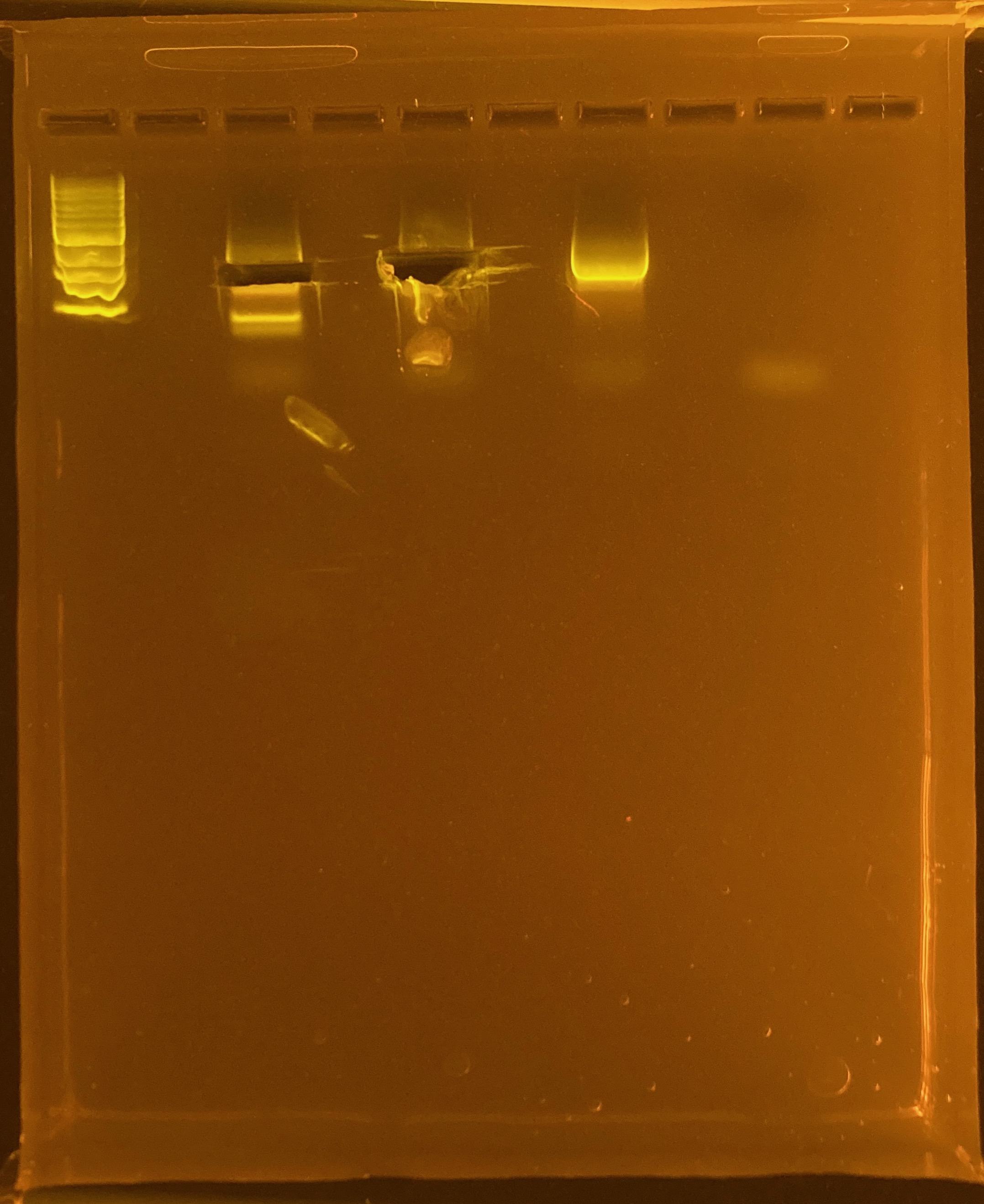
Amplification of 30 POC samples with mtORF on 4/11/2023.
| n. | timepoint | collection_site | colony_id | Extraction Date | Tech | Primerset | PCR Date |
|---|---|---|---|---|---|---|---|
| 81 | TP1 | site2 | POC-217 | 20211015 | KXT | mtORF | 4/11/2023 |
| 267 | TP2 | site2 | POC-257 | 20211112 | KXT | mtORF | 4/11/2023 |
| 273 | TP2 | site2 | POC-200 | 20211007 | KXT | mtORF | 4/11/2023 |
| 289 | TP2 | site2 | POC-205 | 20210924 | KXT | mtORF | 4/11/2023 |
| 299 | TP2 | site2 | POC-255 | 20211004 | KXT | mtORF | 4/11/2023 |
| 305 | TP2 | site2 | POC-222 | 20210923 | KXT | mtORF | 4/11/2023 |
| 309 | TP2 | site2 | POC-207 | 20211105 | KXT | mtORF | 4/11/2023 |
| 311 | TP2 | site2 | POC-254 | 20211019 | KXT | mtORF | 4/11/2023 |
| 315 | TP2 | site2 | POC-219 | 20210910 | KXT | mtORF | 4/11/2023 |
| 317 | TP2 | site2 | POC-239 | 20211104 | KXT | mtORF | 4/11/2023 |
| 321 | TP2 | site2 | POC-238 | 20211018 | KXT | mtORF | 4/11/2023 |
| 323 | TP2 | site2 | POC-259 | 20211108 | KXT | mtORF | 4/11/2023 |
| 325 | TP2 | site2 | POC-248 | 20211007 | KXT | mtORF | 4/11/2023 |
| 329 | TP2 | site2 | POC-201 | 20210930 | KXT | mtORF | 4/11/2023 |
| 335 | TP2 | site2 | POC-215 | 20211115 | KXT | mtORF | 4/11/2023 |
| 345 | TP2 | site3 | POC-371 | 20210907 | KXT | mtORF | 4/11/2023 |
| 349 | TP2 | site3 | POC-372 | 20210831 | KXT | mtORF | 4/11/2023 |
| 355 | TP2 | site3 | POC-373 | 20210920 | KXT | mtORF | 4/11/2023 |
| 363 | TP2 | site3 | POC-366 | 20210916 | KXT | mtORF | 4/11/2023 |
| 367 | TP2 | site3 | POC-391 | 20211109 | KXT | mtORF | 4/11/2023 |
| 369 | TP2 | site3 | POC-394 | 20210927 | KXT | mtORF | 4/11/2023 |
| 373 | TP2 | site3 | POC-359 | 20211108 | KXT | mtORF | 4/11/2023 |
| 377 | TP2 | site3 | POC-346 | 20211115 | KXT | mtORF | 4/11/2023 |
| 419 | TP2 | site3 | POC-369 | 20211101 | KXT | mtORF | 4/11/2023 |
| 437 | TP2 | site3 | POC-358 | 20211014 | KXT | mtORF | 4/11/2023 |
| 447 | TP2 | site3 | POC-395 | 20211122 | KXT | mtORF | 4/11/2023 |
| 449 | TP2 | site3 | POC-377 | 20211129 | KXT | mtORF | 4/11/2023 |
| 455 | TP2 | site3 | POC-375 | 20210921 | KXT | mtORF | 4/11/2023 |
| 457 | TP2 | site3 | POC-378 | 20211008 | KXT | mtORF | 4/11/2023 |
| 465 | TP2 | site3 | POC-386 | 20211005 | KXT | mtORF | 4/11/2023 |
Positive control: Kristina’s E5 extracted DNA Tube #383 (POC 45).
DNA and RNA Quality Check: 1% Gel at 80V for 30 mins.
Qubit Values:
| n. | Qubit AVG | Nanodrop |
|---|---|---|
| 81 | 17.65 | 121.00 |
| 267 | 17.20 | 125.90 |
| 273 | 17.00 | 131.40 |
| 289 | 15.60 | 118.60 |
| 299 | 16.15 | 123.00 |
| 305 | 16.40 | 119.00 |
| 309 | 16.15 | 117.90 |
| 311 | 16.90 | 115.40 |
| 315 | 19.65 | 129.10 |
| 317 | 16.70 | 134.80 |
| 321 | 17.10 | 132.20 |
| 323 | 16.30 | 125.40 |
| 325 | 18.25 | 132.20 |
| 329 | 18.15 | 132.40 |
| 335 | 19.55 | 143.70 |
| 345 | 15.20 | 126.50 |
| 349 | 15.90 | 128.90 |
| 355 | 14.00 | 135.80 |
| 363 | 17.50 | 126.70 |
| 367 | 13.55 | 128.90 |
| 369 | 17.50 | 131.20 |
| 373 | 17.50 | 130.90 |
| 377 | 14.15 | 99.20 |
| 419 | 18.95 | 119.60 |
| 437 | 11.85 | 88.00 |
| 447 | 11.85 | 96.90 |
| 449 | 17.40 | 117.50 |
| 455 | 13.90 | 105.90 |
| 457 | 13.35 | 106.90 |
| 465 | 13.15 | 92.80 |
SET UP for sequencing USING QUBIT VALUES (did not have access to nanodrop at the time, went back later and got nanodrop values), so did not dilute the PCR product.
| Sample ID | Well | Template | A. Template size | B. Template stock conc. (ng/ul) | C. PCR template: ng needed | D. Volume=(C/B)ul | E | F. Volume PCR H20 needed (10-D or E) | G. Primer Vol | Sample # | Colony |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PCR | 1000 | 17.65 | 25 | 1.4 | 8.6 | 2 | 81 | POC-217 | ||
| 2 | PCR | 1000 | 17.65 | 25 | 1.4 | 8.6 | 2 | 81 | POC-217 | ||
| 3 | PCR | 1000 | 17.20 | 25 | 1.5 | 8.5 | 2 | 267 | POC-257 | ||
| 4 | PCR | 1000 | 17.20 | 25 | 1.5 | 8.5 | 2 | 267 | POC-257 | ||
| 5 | PCR | 1000 | 17.00 | 25 | 1.5 | 8.5 | 2 | 273 | POC-200 | ||
| 6 | PCR | 1000 | 17.00 | 25 | 1.5 | 8.5 | 2 | 273 | POC-200 | ||
| 7 | PCR | 1000 | 15.60 | 25 | 1.6 | 8.4 | 2 | 289 | POC-205 | ||
| 8 | PCR | 1000 | 15.60 | 25 | 1.6 | 8.4 | 2 | 289 | POC-205 | ||
| 9 | PCR | 1000 | 16.15 | 25 | 1.5 | 8.5 | 2 | 299 | POC-255 | ||
| 10 | PCR | 1000 | 16.15 | 25 | 1.5 | 8.5 | 2 | 299 | POC-255 | ||
| 11 | PCR | 1000 | 16.40 | 25 | 1.5 | 8.5 | 2 | 305 | POC-222 | ||
| 12 | PCR | 1000 | 16.40 | 25 | 1.5 | 8.5 | 2 | 305 | POC-222 | ||
| 13 | PCR | 1000 | 16.15 | 25 | 1.5 | 8.5 | 2 | 309 | POC-207 | ||
| 14 | PCR | 1000 | 16.15 | 25 | 1.5 | 8.5 | 2 | 309 | POC-207 | ||
| 15 | PCR | 1000 | 16.90 | 25 | 1.5 | 8.5 | 2 | 311 | POC-254 | ||
| 16 | PCR | 1000 | 16.90 | 25 | 1.5 | 8.5 | 2 | 311 | POC-254 | ||
| 17 | PCR | 1000 | 19.65 | 25 | 1.3 | 8.7 | 2 | 315 | POC-219 | ||
| 18 | PCR | 1000 | 19.65 | 25 | 1.3 | 8.7 | 2 | 315 | POC-219 | ||
| 19 | PCR | 1000 | 16.70 | 25 | 1.5 | 8.5 | 2 | 317 | POC-239 | ||
| 20 | PCR | 1000 | 16.70 | 25 | 1.5 | 8.5 | 2 | 317 | POC-239 | ||
| 21 | PCR | 1000 | 17.10 | 25 | 1.5 | 8.5 | 2 | 321 | POC-238 | ||
| 22 | PCR | 1000 | 17.10 | 25 | 1.5 | 8.5 | 2 | 321 | POC-238 | ||
| 23 | PCR | 1000 | 16.30 | 25 | 1.5 | 8.5 | 2 | 323 | POC-259 | ||
| 24 | PCR | 1000 | 16.30 | 25 | 1.5 | 8.5 | 2 | 323 | POC-259 | ||
| 25 | PCR | 1000 | 18.25 | 25 | 1.4 | 8.6 | 2 | 325 | POC-248 | ||
| 26 | PCR | 1000 | 18.25 | 25 | 1.4 | 8.6 | 2 | 325 | POC-248 | ||
| 27 | PCR | 1000 | 18.15 | 25 | 1.4 | 8.6 | 2 | 329 | POC-201 | ||
| 28 | PCR | 1000 | 18.15 | 25 | 1.4 | 8.6 | 2 | 329 | POC-201 | ||
| 29 | PCR | 1000 | 19.55 | 25 | 1.3 | 8.7 | 2 | 335 | POC-215 | ||
| 30 | PCR | 1000 | 19.55 | 25 | 1.3 | 8.7 | 2 | 335 | POC-215 | ||
| 31 | PCR | 1000 | 15.20 | 25 | 1.6 | 8.4 | 2 | 345 | POC-371 | ||
| 32 | PCR | 1000 | 15.20 | 25 | 1.6 | 8.4 | 2 | 345 | POC-371 | ||
| 33 | PCR | 1000 | 15.90 | 25 | 1.6 | 8.4 | 2 | 349 | POC-372 | ||
| 34 | PCR | 1000 | 15.90 | 25 | 1.6 | 8.4 | 2 | 349 | POC-372 | ||
| 35 | PCR | 1000 | 14.00 | 25 | 1.8 | 8.2 | 2 | 355 | POC-373 | ||
| 36 | PCR | 1000 | 14.00 | 25 | 1.8 | 8.2 | 2 | 355 | POC-373 | ||
| 37 | PCR | 1000 | 17.50 | 25 | 1.4 | 8.6 | 2 | 363 | POC-366 | ||
| 38 | PCR | 1000 | 17.50 | 25 | 1.4 | 8.6 | 2 | 363 | POC-366 | ||
| 39 | PCR | 1000 | 13.55 | 25 | 1.8 | 8.2 | 2 | 367 | POC-391 | ||
| 40 | PCR | 1000 | 13.55 | 25 | 1.8 | 8.2 | 2 | 367 | POC-391 | ||
| 41 | PCR | 1000 | 17.50 | 25 | 1.4 | 8.6 | 2 | 369 | POC-394 | ||
| 42 | PCR | 1000 | 17.50 | 25 | 1.4 | 8.6 | 2 | 369 | POC-394 | ||
| 43 | PCR | 1000 | 17.50 | 25 | 1.4 | 8.6 | 2 | 373 | POC-359 | ||
| 44 | PCR | 1000 | 17.50 | 25 | 1.4 | 8.6 | 2 | 373 | POC-359 | ||
| 45 | PCR | 1000 | 14.15 | 25 | 1.8 | 8.2 | 2 | 377 | POC-346 | ||
| 46 | PCR | 1000 | 14.15 | 25 | 1.8 | 8.2 | 2 | 377 | POC-346 | ||
| 47 | PCR | 1000 | 18.95 | 25 | 1.3 | 8.7 | 2 | 419 | POC-369 | ||
| 48 | PCR | 1000 | 18.95 | 25 | 1.3 | 8.7 | 2 | 419 | POC-369 | ||
| 49 | PCR | 1000 | 11.85 | 25 | 2.1 | 7.9 | 2 | 437 | POC-358 | ||
| 50 | PCR | 1000 | 11.85 | 25 | 2.1 | 7.9 | 2 | 437 | POC-358 | ||
| 51 | PCR | 1000 | 11.85 | 25 | 2.1 | 7.9 | 2 | 447 | POC-395 | ||
| 52 | PCR | 1000 | 11.85 | 25 | 2.1 | 7.9 | 2 | 447 | POC-395 | ||
| 53 | PCR | 1000 | 17.40 | 25 | 1.4 | 8.6 | 2 | 449 | POC-377 | ||
| 54 | PCR | 1000 | 17.40 | 25 | 1.4 | 8.6 | 2 | 449 | POC-377 | ||
| 55 | PCR | 1000 | 13.90 | 25 | 1.8 | 8.2 | 2 | 455 | POC-375 | ||
| 56 | PCR | 1000 | 13.90 | 25 | 1.8 | 8.2 | 2 | 455 | POC-375 | ||
| 57 | PCR | 1000 | 13.35 | 25 | 1.9 | 8.1 | 2 | 457 | POC-378 | ||
| 58 | PCR | 1000 | 13.35 | 25 | 1.9 | 8.1 | 2 | 457 | POC-378 | ||
| 59 | PCR | 1000 | 13.15 | 25 | 1.9 | 8.1 | 2 | 465 | POC-386 | ||
| 60 | PCR | 1000 | 13.15 | 25 | 1.9 | 8.1 | 2 | 465 | POC-386 |
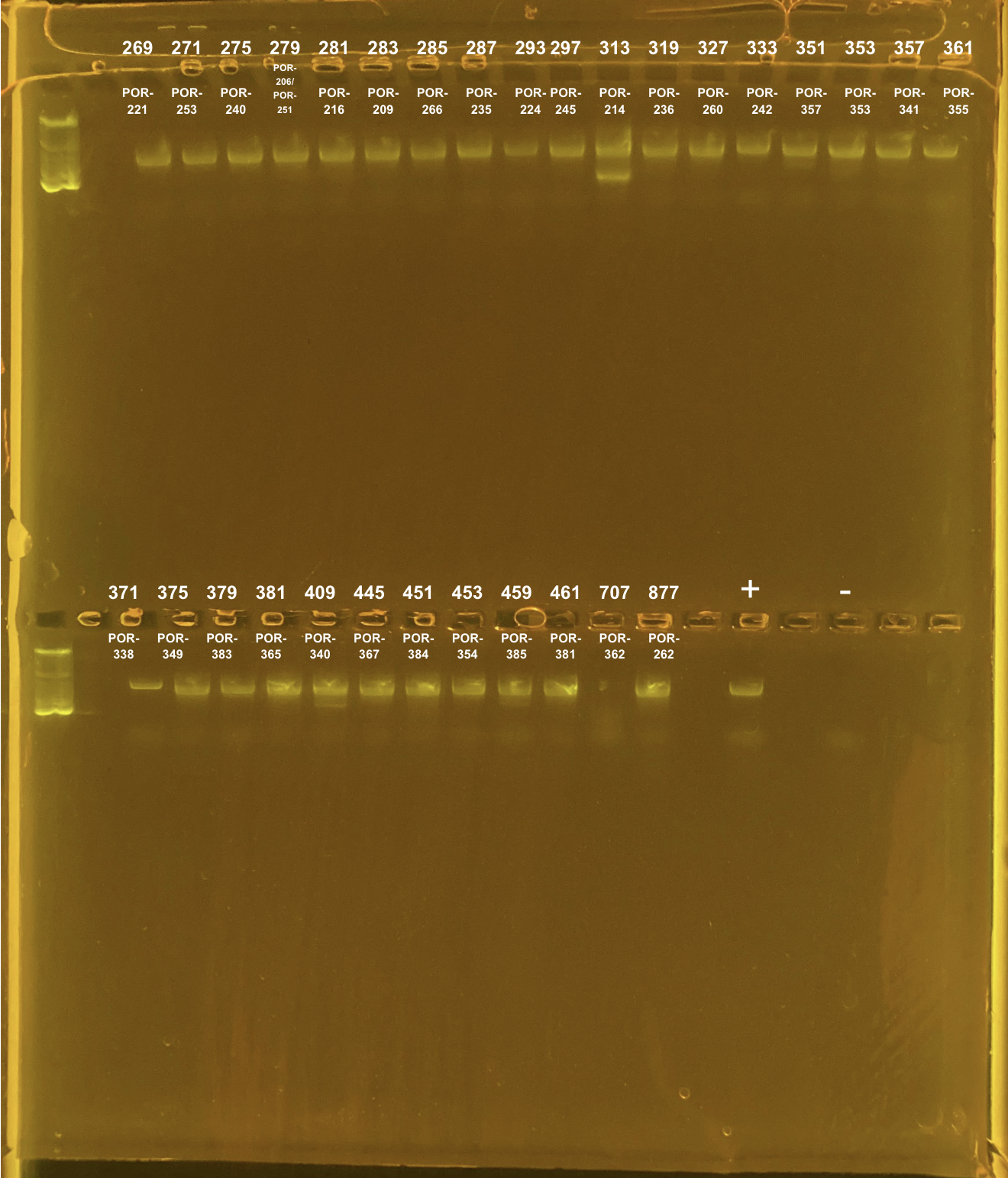
Amplification of 30 POR samples on 4/13/2023.
| n. | timepoint | collection_site | colony_id | Extraction Date | Tech | Primerset | PCR Date |
|---|---|---|---|---|---|---|---|
| 269 | TP2 | site2 | POR-221 | 20211102 | KXT | H2 | 4/13/2023 |
| 271 | TP2 | site2 | POR-253 | 20211112 | KXT | H2 | 4/13/2023 |
| 275 | TP2 | site2 | POR-240 | 20211007 | KXT | H2 | 4/13/2023 |
| 279 | TP2 | site2 | POR-206/POR-251 | 20211115 | KXT | H2 | 4/13/2023 |
| 281 | TP2 | site2 | POR-216 | 20211004 | KXT | H2 | 4/13/2023 |
| 283 | TP2 | site2 | POR-209 | 20211108 | KXT | H2 | 4/13/2023 |
| 285 | TP2 | site2 | POR-266 | 20211112 | KXT | H2 | 4/13/2023 |
| 287 | TP2 | site2 | POR-235 | 20220201 | KXT | H2 | 4/13/2023 |
| 293 | TP2 | site2 | POR-224 | 20211105 | KXT | H2 | 4/13/2023 |
| 297 | TP2 | site2 | POR-245 | 20210924 | KXT | H2 | 4/13/2023 |
| 313 | TP2 | site2 | POR-214 | 20210907 | KXT | H2 | 4/13/2023 |
| 319 | TP2 | site2 | POR-236 | 20210923 | KXT | H2 | 4/13/2023 |
| 327 | TP2 | site2 | POR-260 | 20210930 | KXT | H2 | 4/13/2023 |
| 333 | TP2 | site2 | POR-242 | 20210913 | KXT | H2 | 4/13/2023 |
| 351 | TP2 | site3 | POR-357 | 20211019 | KXT | H2 | 4/13/2023 |
| 353 | TP2 | site3 | POR-353 | 20210831 | KXT | H2 | 4/13/2023 |
| 357 | TP2 | site3 | POR-341 | 20210916 | KXT | H2 | 4/13/2023 |
| 361 | TP2 | site3 | POR-355 | 20210913 | KXT | H2 | 4/13/2023 |
| 371 | TP2 | site3 | POR-338 | 20210913 | KXT | H2 | 4/13/2023 |
| 375 | TP2 | site3 | POR-349 | 20210920 | KXT | H2 | 4/13/2023 |
| 379 | TP2 | site3 | POR-383 | 20211012 | KXT | H2 | 4/13/2023 |
| 381 | TP2 | site3 | POR-365 | 20211001 | KXT | H2 | 4/13/2023 |
| 409 | TP2 | site3 | POR-340 | 20210903 | KXT | H2 | 4/13/2023 |
| 445 | TP2 | site3 | POR-367 | 20211018 | KXT | H2 | 4/13/2023 |
| 451 | TP2 | site3 | POR-384 | 20211005 | KXT | H2 | 4/13/2023 |
| 453 | TP2 | site3 | POR-354 | 20211028 | KXT | H2 | 4/13/2023 |
| 459 | TP2 | site3 | POR-385 | 20220208 | KXT | H2 | 4/13/2023 |
| 461 | TP2 | site3 | POR-381 | 20211118 | KXT | H2 | 4/13/2023 |
| 707 | TP3 | site3 | POR-362 | 20211020 | KXT | H2 | 4/13/2023 |
| 877 | TP4 | site2 | POR-262 | 20211018 | KXT | H2 | 4/13/2023 |
Positive control: Kristina’s E5 extracted DNA Tube #469 (extraction date: 20220208, PCR’d by me successfully in this post).
DNA and RNA Quality Check: 1% Gel at 80V for 30 mins.
Did not clean up the double banding samples:
313 TP2 site2 POR-214
409 TP2 site3 POR-340
Nanodrop values
| n. | Nanodrop |
|---|---|
| 269 | 99.20 |
| 271 | 103.80 |
| 275 | 141.50 |
| 279 | 155.50 |
| 281 | 143.70 |
| 283 | 146.90 |
| 285 | 138.60 |
| 287 | 99.90 |
| 293 | 113.50 |
| 297 | 154.30 |
| 313 | Double banded |
| 319 | 149.20 |
| 327 | 109.80 |
| 333 | 110.30 |
| 351 | 129.80 |
| 353 | 153.30 |
| 357 | 129.90 |
| 361 | 133.40 |
| 371 | 95.40 |
| 375 | 135.50 |
| 379 | 130.00 |
| 381 | 153.30 |
| 409 | Double banded |
| 445 | 144.70 |
| 451 | 122.80 |
| 453 | 106.70 |
| 459 | 114.50 |
| 461 | 124.70 |
| 707 | 61.80 |
| 877 | 115.90 |
SET UP the 28 that did not have double banding for sequencing 1:10th dilutions from nanodrop concentration
| Sample ID | Well | Template | A. Template size | B. Template stock conc. (ng/ul) | C. PCR template: ng needed | D. Volume=(C/B)ul | E | F. Volume PCR H20 needed (10-D or E) | G. Primer Vol | Sample # | Colony | Undiluted nanodrop concentration |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 69 | PCR | 1500 | 9.92 | 37.5 | 3.8 | 6.2 | 2 | 269 | POR-221 | 99.20 | ||
| 70 | PCR | 1500 | 9.92 | 37.5 | 3.8 | 6.2 | 2 | 269 | POR-221 | 99.20 | ||
| 71 | PCR | 1500 | 10.38 | 37.5 | 3.6 | 6.4 | 2 | 271 | POR-253 | 103.80 | ||
| 72 | PCR | 1500 | 10.38 | 37.5 | 3.6 | 6.4 | 2 | 271 | POR-253 | 103.80 | ||
| 73 | PCR | 1500 | 14.15 | 37.5 | 2.7 | 7.3 | 2 | 275 | POR-240 | 141.50 | ||
| 74 | PCR | 1500 | 14.15 | 37.5 | 2.7 | 7.3 | 2 | 275 | POR-240 | 141.50 | ||
| 75 | PCR | 1500 | 15.55 | 37.5 | 2.4 | 7.6 | 2 | 279 | POR-206/POR-251 | 155.50 | ||
| 76 | PCR | 1500 | 15.55 | 37.5 | 2.4 | 7.6 | 2 | 279 | POR-206/POR-251 | 155.50 | ||
| 77 | PCR | 1500 | 14.37 | 37.5 | 2.6 | 7.4 | 2 | 281 | POR-216 | 143.70 | ||
| 78 | PCR | 1500 | 14.37 | 37.5 | 2.6 | 7.4 | 2 | 281 | POR-216 | 143.70 | ||
| 79 | PCR | 1500 | 14.69 | 37.5 | 2.6 | 7.4 | 2 | 283 | POR-209 | 146.90 | ||
| 80 | PCR | 1500 | 14.69 | 37.5 | 2.6 | 7.4 | 2 | 283 | POR-209 | 146.90 | ||
| 81 | PCR | 1500 | 13.86 | 37.5 | 2.7 | 7.3 | 2 | 285 | POR-266 | 138.60 | ||
| 82 | PCR | 1500 | 13.86 | 37.5 | 2.7 | 7.3 | 2 | 285 | POR-266 | 138.60 | ||
| 83 | PCR | 1500 | 9.99 | 37.5 | 3.8 | 6.2 | 2 | 287 | POR-235 | 99.90 | ||
| 84 | PCR | 1500 | 9.99 | 37.5 | 3.8 | 6.2 | 2 | 287 | POR-235 | 99.90 | ||
| 85 | PCR | 1500 | 11.35 | 37.5 | 3.3 | 6.7 | 2 | 293 | POR-224 | 113.50 | ||
| 86 | PCR | 1500 | 11.35 | 37.5 | 3.3 | 6.7 | 2 | 293 | POR-224 | 113.50 | ||
| 87 | PCR | 1500 | 15.43 | 37.5 | 2.4 | 7.6 | 2 | 297 | POR-245 | 154.30 | ||
| 88 | PCR | 1500 | 15.43 | 37.5 | 2.4 | 7.6 | 2 | 297 | POR-245 | 154.30 | ||
| 89 | PCR | 1500 | 14.92 | 37.5 | 2.5 | 7.5 | 2 | 319 | POR-236 | 149.20 | ||
| 90 | PCR | 1500 | 14.92 | 37.5 | 2.5 | 7.5 | 2 | 319 | POR-236 | 149.20 | ||
| 91 | PCR | 1500 | 10.98 | 37.5 | 3.4 | 6.6 | 2 | 327 | POR-260 | 109.80 | ||
| 92 | PCR | 1500 | 10.98 | 37.5 | 3.4 | 6.6 | 2 | 327 | POR-260 | 109.80 | ||
| 93 | PCR | 1500 | 11.03 | 37.5 | 3.4 | 6.6 | 2 | 333 | POR-242 | 110.30 | ||
| 94 | PCR | 1500 | 11.03 | 37.5 | 3.4 | 6.6 | 2 | 333 | POR-242 | 110.30 | ||
| 95 | PCR | 1500 | 12.98 | 37.5 | 2.9 | 7.1 | 2 | 351 | POR-357 | 129.80 | ||
| 96 | PCR | 1500 | 12.98 | 37.5 | 2.9 | 7.1 | 2 | 351 | POR-357 | 129.80 | ||
| 97 | PCR | 1500 | 15.33 | 37.5 | 2.4 | 7.6 | 2 | 353 | POR-353 | 153.30 | ||
| 98 | PCR | 1500 | 15.33 | 37.5 | 2.4 | 7.6 | 2 | 353 | POR-353 | 153.30 | ||
| 99 | PCR | 1500 | 12.99 | 37.5 | 2.9 | 7.1 | 2 | 357 | POR-341 | 129.90 | ||
| 100 | PCR | 1500 | 12.99 | 37.5 | 2.9 | 7.1 | 2 | 357 | POR-341 | 129.90 | ||
| 101 | PCR | 1500 | 13.34 | 37.5 | 2.8 | 7.2 | 2 | 361 | POR-355 | 133.40 | ||
| 102 | PCR | 1500 | 13.34 | 37.5 | 2.8 | 7.2 | 2 | 361 | POR-355 | 133.40 | ||
| 103 | PCR | 1500 | 9.54 | 37.5 | 3.9 | 6.1 | 2 | 371 | POR-338 | 95.40 | ||
| 104 | PCR | 1500 | 9.54 | 37.5 | 3.9 | 6.1 | 2 | 371 | POR-338 | 95.40 | ||
| 105 | PCR | 1500 | 13.55 | 37.5 | 2.8 | 7.2 | 2 | 375 | POR-349 | 135.50 | ||
| 106 | PCR | 1500 | 13.55 | 37.5 | 2.8 | 7.2 | 2 | 375 | POR-349 | 135.50 | ||
| 107 | PCR | 1500 | 13.00 | 37.5 | 2.9 | 7.1 | 2 | 379 | POR-383 | 130.00 | ||
| 108 | PCR | 1500 | 13.00 | 37.5 | 2.9 | 7.1 | 2 | 379 | POR-383 | 130.00 | ||
| 109 | PCR | 1500 | 15.33 | 37.5 | 2.4 | 7.6 | 2 | 381 | POR-365 | 153.30 | ||
| 110 | PCR | 1500 | 15.33 | 37.5 | 2.4 | 7.6 | 2 | 381 | POR-365 | 153.30 | ||
| 111 | PCR | 1500 | 14.47 | 37.5 | 2.6 | 7.4 | 2 | 445 | POR-367 | 144.70 | ||
| 112 | PCR | 1500 | 14.47 | 37.5 | 2.6 | 7.4 | 2 | 445 | POR-367 | 144.70 | ||
| 113 | PCR | 1500 | 12.28 | 37.5 | 3.1 | 6.9 | 2 | 451 | POR-384 | 122.80 | ||
| 114 | PCR | 1500 | 12.28 | 37.5 | 3.1 | 6.9 | 2 | 451 | POR-384 | 122.80 | ||
| 115 | PCR | 1500 | 10.67 | 37.5 | 3.5 | 6.5 | 2 | 453 | POR-354 | 106.70 | ||
| 116 | PCR | 1500 | 10.67 | 37.5 | 3.5 | 6.5 | 2 | 453 | POR-354 | 106.70 | ||
| 117 | PCR | 1500 | 11.45 | 37.5 | 3.3 | 6.7 | 2 | 459 | POR-385 | 114.50 | ||
| 118 | PCR | 1500 | 11.45 | 37.5 | 3.3 | 6.7 | 2 | 459 | POR-385 | 114.50 | ||
| 119 | PCR | 1500 | 12.47 | 37.5 | 3.0 | 7.0 | 2 | 461 | POR-381 | 124.70 | ||
| 120 | PCR | 1500 | 12.47 | 37.5 | 3.0 | 7.0 | 2 | 461 | POR-381 | 124.70 | ||
| 121 | PCR | 1500 | 6.18 | 37.5 | 6.1 | 3.9 | 2 | 707 | POR-362 | 61.80 | ||
| 122 | PCR | 1500 | 6.18 | 37.5 | 6.1 | 3.9 | 2 | 707 | POR-362 | 61.80 | ||
| 123 | PCR | 1500 | 11.59 | 37.5 | 3.2 | 6.8 | 2 | 877 | POR-262 | 115.90 | ||
| 124 | PCR | 1500 | 11.59 | 37.5 | 3.2 | 6.8 | 2 | 877 | POR-262 | 115.90 |
POC mtORF (ZD01-8) test samples were sent For Sanger Sequencing on 4/13/2023. The rest of the POC mtORF (ZD09-68) and POR H2 (ZD69-ZD124) (except for the 2 samples with double banding) will be sent for sequencing on 4/18/2023.
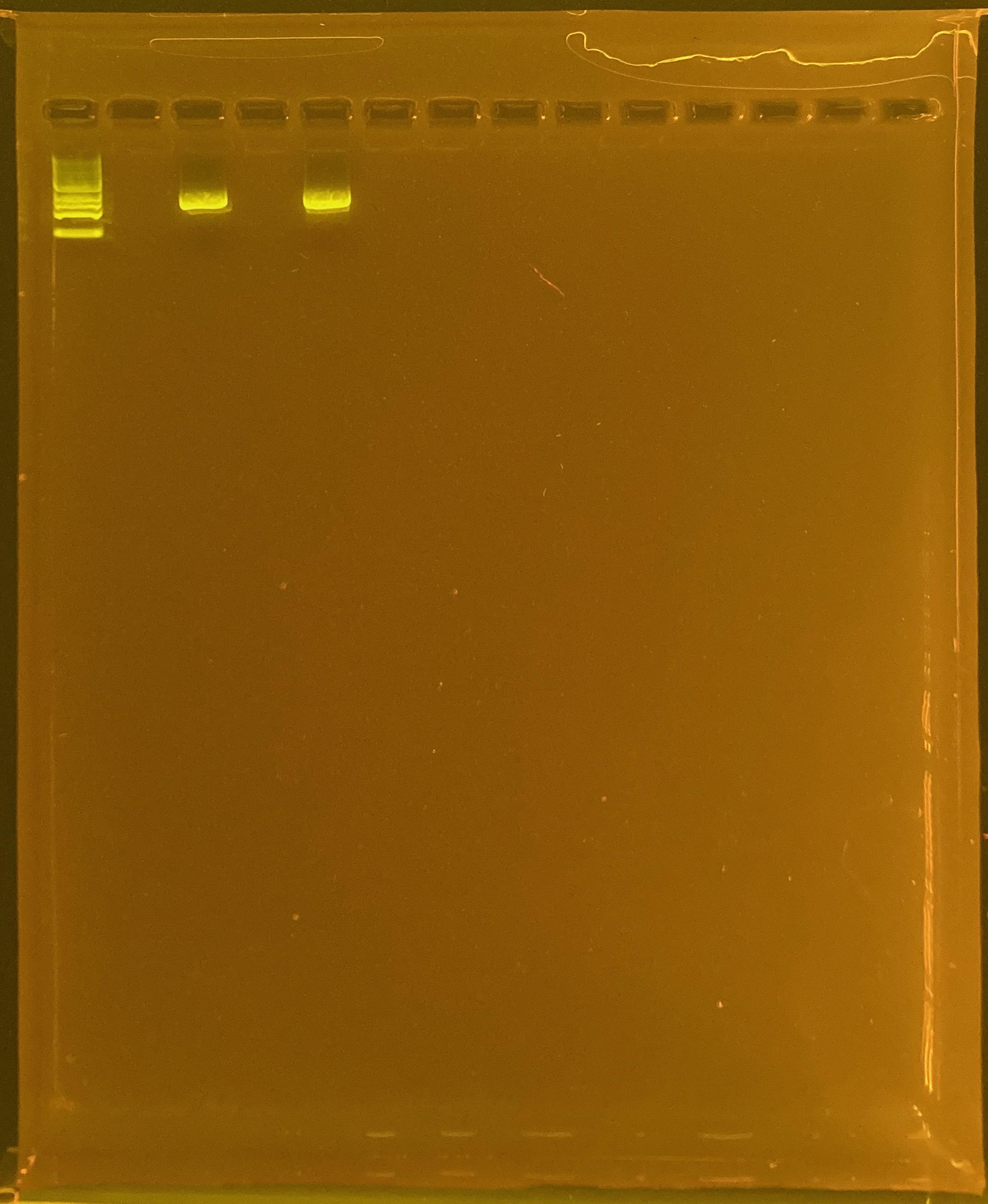
Double Banding in POR H2
These samples had double banding in the gel on 4/13/23
313 TP2 site2 POR-214
409 TP2 site3 POR-340
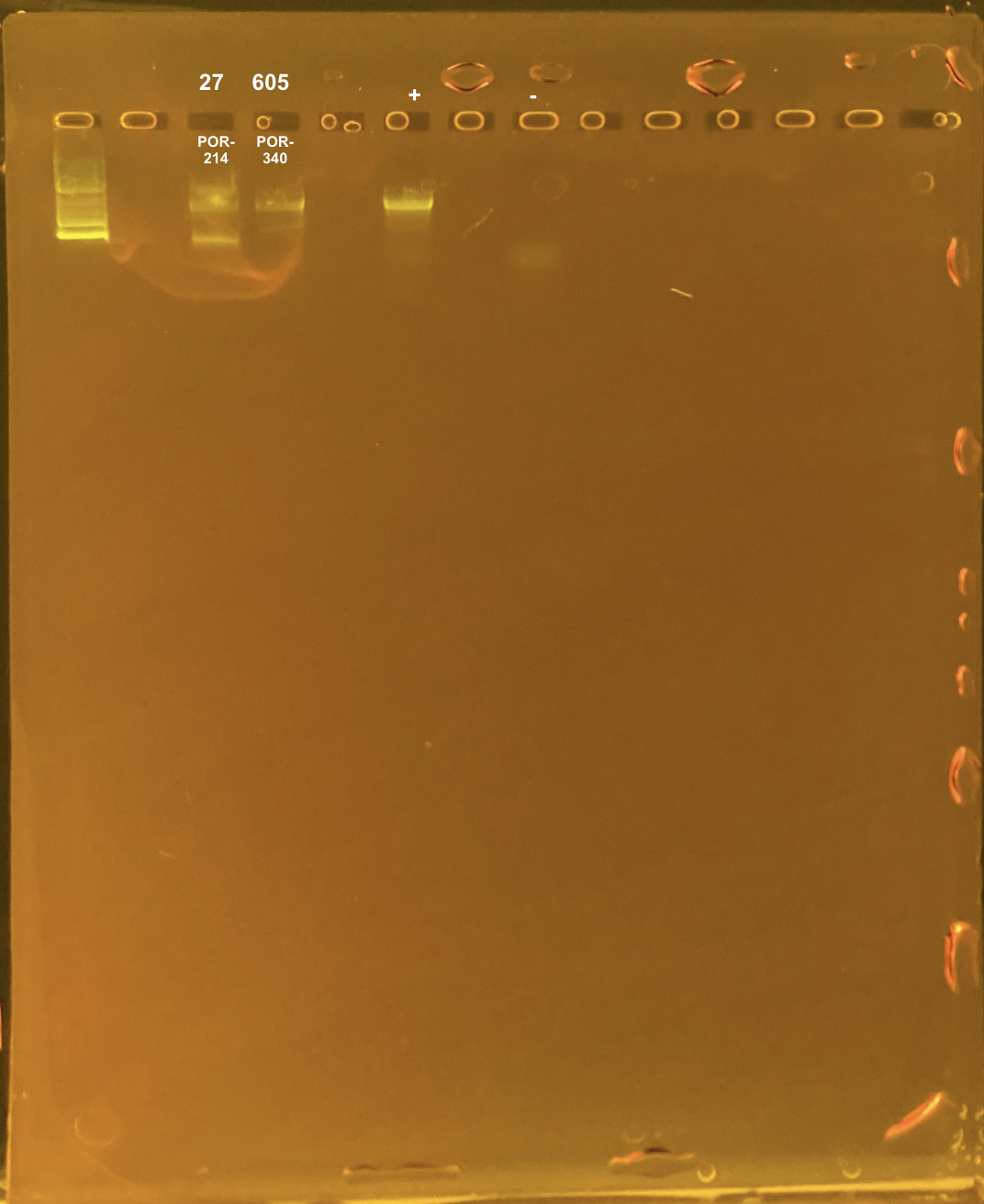
To test if the double banding was due to poor DNA quality, I re-PCRd these colonies from different time points:
27 TP1 site2 POR-214
605 TP3 site3 POR-340
Positive control: Kristina’s E5 extracted DNA Tube #371 (extraction date: 20210913, PCR’d by me successfully above).
DNA and RNA Quality Check: 1% Gel at 80V for 30 mins.
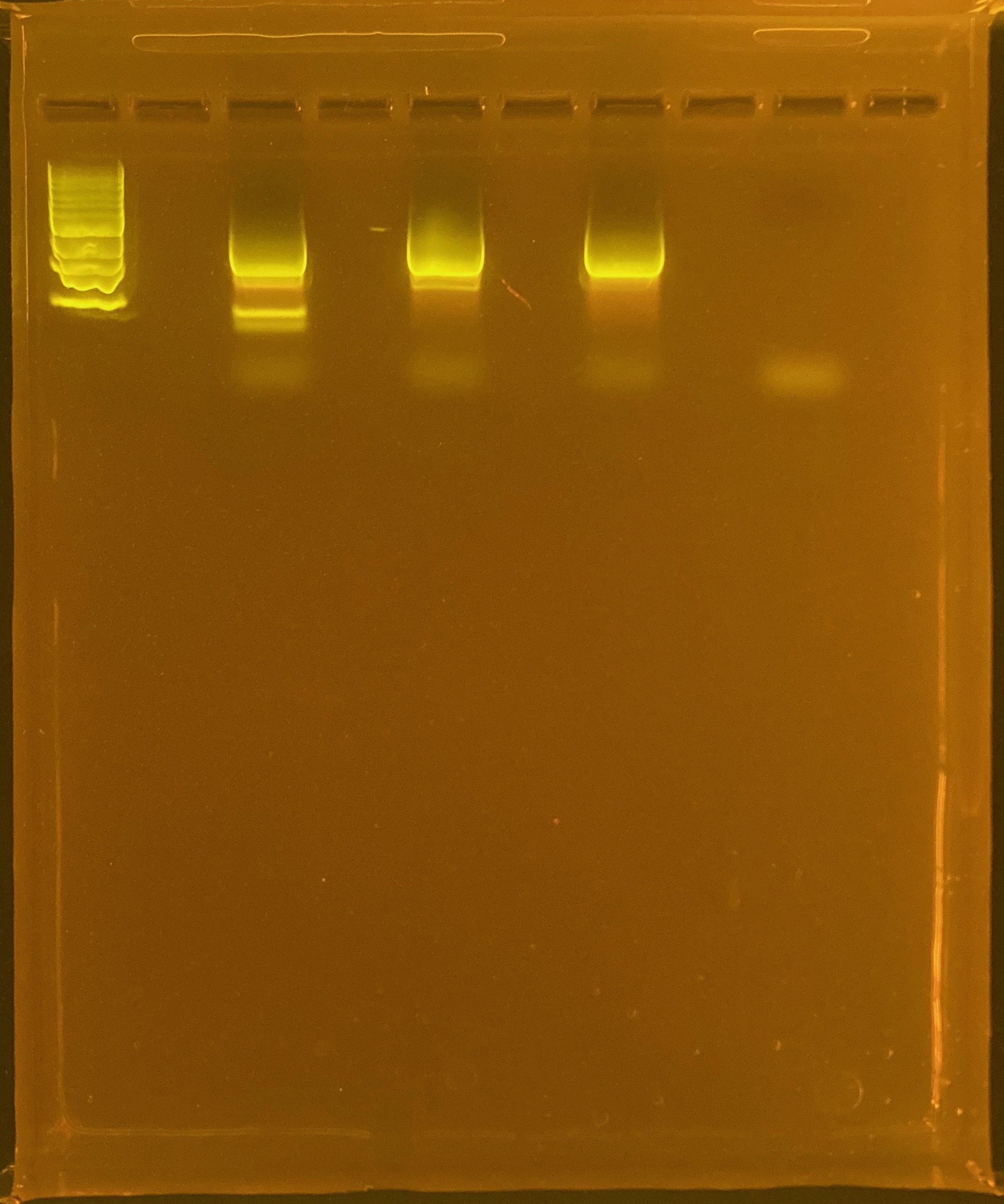
Next steps: run large comb gel with the rest of the PCR product and perform a gell extraction on the band that has the same size as the positive control ~1500bp. Need UV protection.
Excise bands at same length as positive control:
Perform cleanup of excised bands using Zymoclean Kit
(Will write this up ASAP, for now see notebook page images here and here)
Run second gel to quality check and confirm only one band is remaining in the cleaned up product.
Set up for sequencing using nanodrop values (no dilution needed), will be sequenced on 4/24/23.
| Sample ID | Well | Template | A. Template size | B. Template stock conc. (ng/ul) | C. PCR template: ng needed | D. Volume=(C/B)ul | E | F. Volume PCR H20 needed (10-D or E) | G. Primer Vol | Sample # | Colony |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 125 | PCR | 1500 | 32.10 | 37.5 | 1.2 | 8.8 | 2 | 27 | POR-214 | ||
| 126 | PCR | 1500 | 32.10 | 37.5 | 1.2 | 8.8 | 2 | 27 | POR-214 | ||
| 127 | PCR | 1500 | 40.40 | 37.5 | 0.9 | 9.1 | 2 | 605 | POR-340 | ||
| 128 | PCR | 1500 | 40.40 | 37.5 | 0.9 | 9.1 | 2 | 605 | POR-340 |