Testing Charm Biotech RNA Extraction on Cryosectioned P. comp and M. cap
Testing Charm Kit on P. comp and M. cap
Continuation from this post
Protocol Link
Modifications to protocol:
- Modifications to Charm Extraction Protocol
- Pre-cool lysis buffer mixture
- Use increased volume of lysis buffer mixture for whole sections
- 100 uL lysis buffer mixture, 200 uL RB7
- Pre-cool forceps and razor blades for transferring tissue to lysis buffer
- Incubate on a shaker–incubator for 15 min at 56°C and 1400 rpm.
- instead of 1 hour at 52 ºC or 3 hours at 60 ºC. Feels like a big jump, but seems like PAXgene isn’t crosslinking RNA based off of their RNA extraction protocols.
- Stain and wash all slides and perform dissections in petri dishes (or 96-well plate lids) cleaned to be RNAse free
02/01/24 - Sectioining new tissue and attempting to extract RNA from cryosections
- Section the Porites and Montipora fixed and embedded here
- sectioning protocol
- NOTE: These were embedded without a sucrose cryoprotection step beforehand
- Honestly, might continue this, as we have seen mulitple cases of degredation after the sucrose step (see here)
- Notes: was having trouble with the inside of the skeletons. Maybe we could somehow fill this up with OCT?
- Playing around with temperature, mostly sectioned at -19 ºC
- Slides dried in cryostat for 5-10 minutes then transferred to falcon tube with silica gel packet on dry ice (2 slides back to back per tube)
- After sectioning, transported slides to Molecular lab and proceeded with RNA extraction within 1 hour
- Slides were kept in falcon tube with silica gel packet on dry ice
- Rinsed slide in 100% Ethanol for 30s on dry ice
- Replaced ethanol with RNAse/DNAse-free water to rinse
- Remove slide to dark background over PCR rack (over dry ice, though be careful to not freeze the water once the volume on the slide is low, if that’s freezing move to regular ice) and use sterile razor blade, cleaned with RNAse cleaner, to scrape off tissue sections into a tube filled with prepared 100 uL RD2 digestion buffer.
- this is double the normal RD2 amount, because I added a lot of tissue
- honestly just used my tiny forceps this time to scoop up tissue, they were cleaned with ethanol and RNAse cleaner
- Then, I followed the protocol as written for FFPE, with a 15 minute digestion at 56 ºC.
- since I used double the amount of lysis buffer, I used double the amount of binding buffer
- still had some debris in the Porites tube when I transerred to the binding tube, but this seemed okay. In the future, spin down and remove any pellet before proceeding or increase digestion time.
Qubit Results
- Used Broad range RNA Qubit Protocol
- All samples read twice, standard only read once
RNA Standards: 370.23 (S1) & 9494.15 (S2)
| colony_id | RNA_QBIT_1 | RNA_QBIT_2 | RNA_QBIT_AVG |
|---|---|---|---|
| Porites | 27.0 | 27.4 | 27.2 |
| Monitpora | 16.2 | 16.2 | 16.2 |
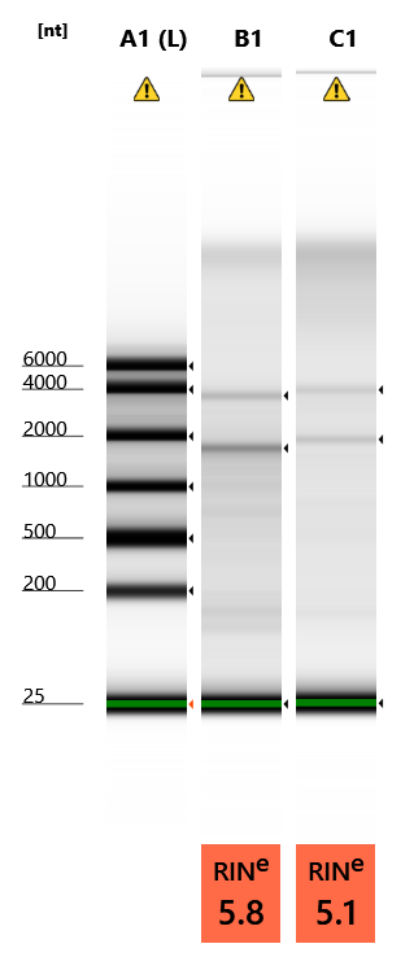
RNA Quality Check: Tapestation
Full results can be found here
Written on February 1, 2024